A research lately revealed in Communications Biology (Nature Portfolio) by Esra Oktay, Farhang Alem, Keziah Hernandez, Michael Girgis, Christopher Inexperienced, Divita Mathur, Igor L. Medintz, Aarthi Narayanan, and Rémí Veneziano introduces an progressive DNA origami–based mostly nanovaccine platform concentrating on the receptor-binding area (RBD) of SARS-CoV-2. Their findings spotlight the potential of rationally engineered DNA nanoparticles to elicit robust and sturdy immune safety.
The researchers designed wireframe DNA origami nanoparticles (DNA-NPs) functionalized with ten trimerized RBD antigens and CpG oligonucleotides as adjuvants. Trimer meeting was achieved by peptide nucleic acid (PNA)–mediated conjugation of protein G to RBD-Fc, enabling exact orientation on DNA-NP overhangs. Biophysical characterization employed dynamic mild scattering, Förster resonance vitality switch (FRET)–based mostly stability assays, and quantitative fluorescence labeling. Structural affirmation was obtained by way of atomic power microscopy (AFM). For prime-resolution AFM imaging, the crew employed NanoWorld® USC-F0.3-k0.3 ultra-short cantilevers, chosen for his or her 300 kHz resonance frequency, ~0.3 N/m power fixed, and 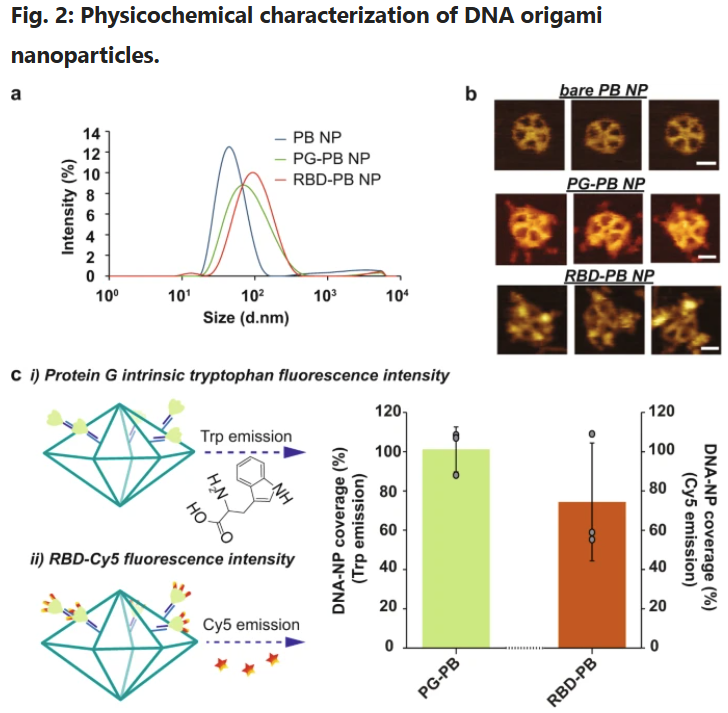
a Hydrodynamic diameter measurement of DNA-NPs with DLS. b Atomic power microscopy imaging of DNA-NPs formation and conjugation with 3-mer RBDs (scale bar: 20 nm). c Fluorescence-intensity based mostly dedication of PG and RBD stoichiometry on DNA origami NP. i) Tryptophan fluorescence emission was used to find out the entire variety of PG loaded on the floor of the DNA-NPs. ii) Second, the stoichiometry of RBD on NP was quantified by way of measuring the emission of Cy5 dyes conjugated to the RBD antigens. The bar graph represents the entire protection share for the PG (inexperienced bar) and the 3-mer RBD (orange bar) on the DNA-NP floor normalized to the variety of conjugation websites out there. Knowledge are proven because the imply ± SD (n = 3 impartial experiments).
This text accommodates photos reused from Jia et al., Nature Communications 14, 1394 (2023), licensed beneath CC BY 4.0.


